OpenCitations Corpus
The OpenCitations Corpus (OCC) is an open repository of scholarly citation data made available under a Creative Commons public domain dedication (CC0), which provides accurate bibliographic references harvested from the scholarly literature that others may freely build upon, enhance and reuse for any purpose, without restriction under copyright or database law. An in-depth description of the OCC is available in the following paper:
Silvio Peroni, David Shotton, Fabio Vitali (2017). One year of the OpenCitations Corpus: Releasing RDF-based scholarly citation data into the Public Domain. In Proceedings of the 16th International Semantic Web Conference (ISWC 2017): 184-192. https://doi.org/10.1007/978-3-319-68204-4_19, OA at https://w3id.org/people/essepuntato/papers/oc-iswc2017.html
The OCC includes information about six different kinds of bibliographic entities:
-
bibliographic resources (br), class
fabio:Expression– resources that either cite or are cited by other bibliographic resources (e.g. journal articles), or that contain such citing/cited resources (e.g. journals); -
resource embodiments (re), class
fabio:Manifestation– details of the physical or digital forms in which the bibliographic resources are made available by their publishers; -
bibliographic references (be), class
biro:BibliographicReference– the literal textual bibliographic references occurring in the reference lists within bibliographic resources, that reference other bibliographic resources; -
responsible agents (ra), class
foaf:Agent– names of agents having certain roles with respect to bibliographic resources (i.e. names of authors, editors, publishers, etc.); -
agent roles (ar), class
pro:RoleInTime– roles held by agents with respect to bibliographic resources (e.g. author, editor, publisher); -
identifiers (id) (class
datacite:Identifier) – external identifiers (e.g. DOI, ORCID, PubMedID) associated with the bibliographic entities.
The corpus URL (https://w3id.org/oc/corpus/) identifies the entire OCC, which is composed of several sub-datasets, one for each of the aforementioned bibliographic entities included in the corpus. Each of these has a URL composed by suffixing the corpus URL with the two-letter short name for the class of entity (e.g. be
for a bibliographic reference) followed by an oblique slash (e.g. https://w3id.org/oc/corpus/be/). Individual members of each sub-dataset are identified by incrementing numbers, unique within that sub-dataset, e.g. https://w3id.org/oc/corpus/br/1 or https://w3id.org/oc/corpus/be/24.
The OCC stores data relevant to these citations in RDF, encoded as JSON-LD, and each dataset is described appropriately by means of the Data Catalog Vocabulary and the VoID Vocabulary.
OCC data are available as dumps on Figshare
The ingestion workflow
The ingestion of citation data into the OCC, briefly summarised in Figure 1, is handled by two Python scripts called the bibliographic references Extractor (BEE) and the SPAR Citation Indexer (SPACIN), available in the OpenCitations GitHub repository.
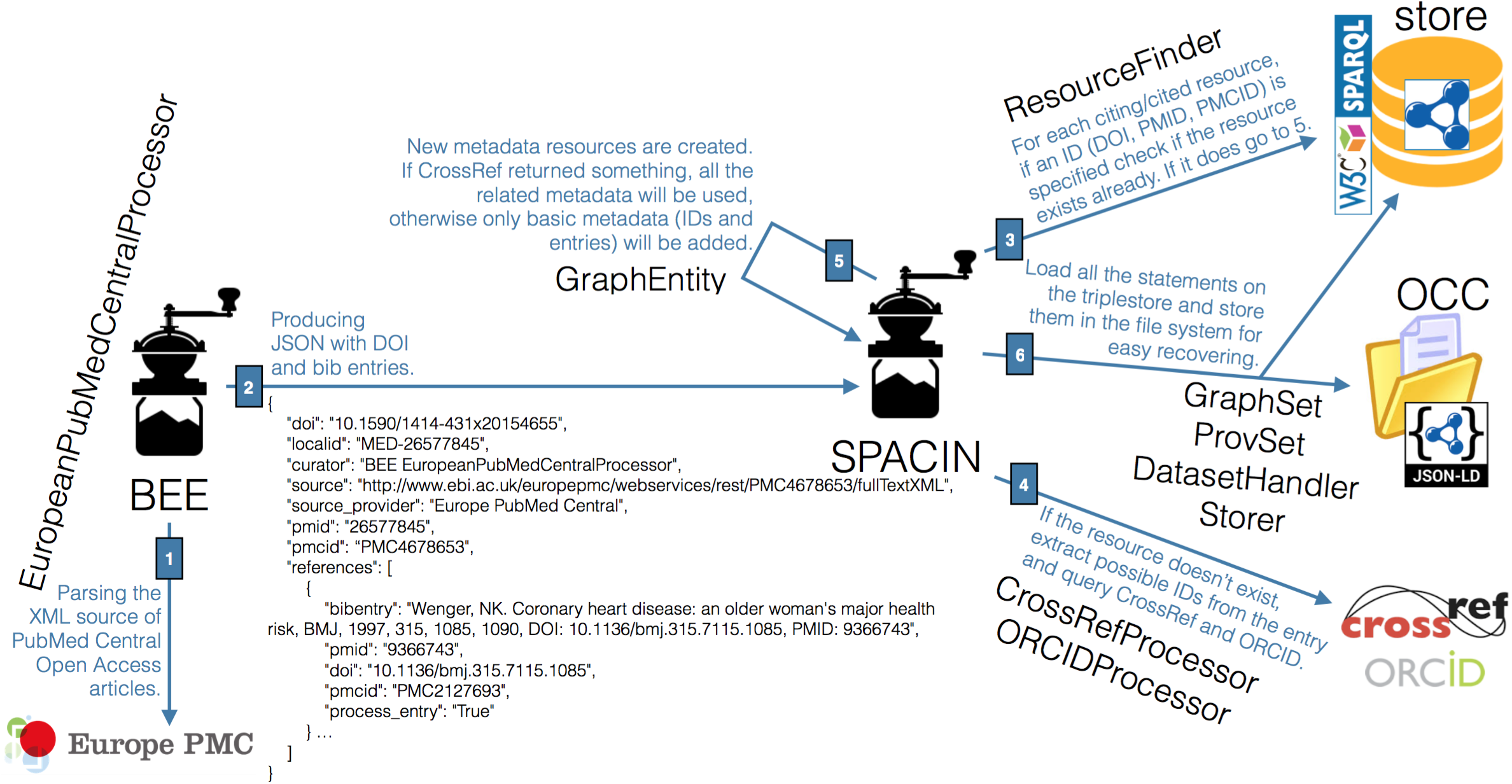
BEE is responsible for the creation of JSON files containing information about the articles in the Open Access subset of PubMed Central (retrieved by using the Europe PubMed Central API). Each of these JSON files is created by asking Europe PubMed Central for all the metadata of the articles it stores, for which the source XML file is available. Once identified, BEE processes all the XML sources so as to extract the complete reference list of the paper under consideration, and includes all these data in the final JSON file. An excerpt of one of those JSON files is introduced as follows:
{
"doi": "10.1007/s11892-016-0752-4",
"pmid": "27168063",
"pmcid": "PMC4863913",
"localid": "MED-27168063",
"curator": "BEE EuropeanPubMedCentralProcessor",
"source": "http://www.ebi.ac.uk/europepmc/webservices/rest/PMC4863913/fullTextXML",
"source_provider": "Europe PubMed Central",
"references": [
...
{
"bibentry": "Chang, KY, Unanue, ER. Prediction of HLA-DQ8beta cell peptidome using a computational program and its relationship to autoreactive T cells, Int Immunol, 2009, 21, 6, 705, 13, DOI: 10.1093/intimm/dxp039, PMID: 19461125",
"pmid": "19461125",
"doi": "10.1093/intimm/dxp039",
"pmcid": "PMC2686615",
"process_entry": "True"
},
...
]
}In particular, for each article retrieved by means of the Europe PubMed Central API, BEE stores all the possible identifiers (in the example, doi
, pmid
, pmcid
, and localid
) and all the textual references, enriched by their own related identifiers if these are available. In addition, the JSON file also includes provenance information about the source, its provider and the curator (i.e. the particular BEE Python class responsible for the extraction of these metadata from the source).
Starting from the output provided by BEE, SPACIN processes each JSON file, retrieving metadata information about all the citing/cited articles described in it by querying the Crossref API and the ORCID API. These APIs are also used to disambiguate bibliographic resources and agents by means of the identifiers retrieved (e.g., DOI, ISSN, ISBN, ORCID, URL, and Crossref Member URL). Once SPACIN has retrieved all these metadata, appropriate RDF resources are created (or reused, if they have been already added to the OCC in the past). These are stored in the file system in JSON-LD format and additionally within the OCC triplestore. It is worth noting that, for space and performance reasons, the triplestore includes all the data about the curated entities, but does not store their provenance data nor the descriptions of the datasets themselves, which are accessible only via HTTP.
The SPACIN workflow introduced in Figure 1 is a process that runs until no more JSON files are available from BEE. Thus, the current instance of the OCC is evolving dynamically in time (even if now it has been stopped for updating the ingestion scripts), and can be easily extended beyond ingest from Europe PubMed Central by reconfiguring it to interact with additional REST APIs provided by different bibliographic sources, so as to gather new article metadata and their related references, thereby expanding the scope and coverage provided by the OCC.
As of March 19, 2022, the OCC has ingested the references from 326743 citing bibliographic resources and contains information about 13964148 citation links to 7565367 cited resources.